Genome Code of HIV
The genome and proteins of HIV (human immunodeficiency virus) have been the subject of extensive research since its discovery in 1983. In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), known to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS, which was later named HIV. Each virion comprises a viral envelope and associated matrix enclosing a capsid that, in turn, contains two copies of the single-stranded RNA genome along with several essential enzymes. The virus was discovered two years after the first major cases of AIDS-associated illnesses were reported.
Structure
Diagram of HIV:
Capsid and Spike Protein: The immature HIV-1 capsid has a cone-shaped core containing the RNA genome and enzymes (reverse transcriptase, integrase, and protease). A limited number of envelope glycoprotein trimers—composed of gp120 and gp41—are embedded in the host-derived membrane, where they mediate binding to the CD4 receptor and co-receptors (CCR5 or CXCR4) on target cells.
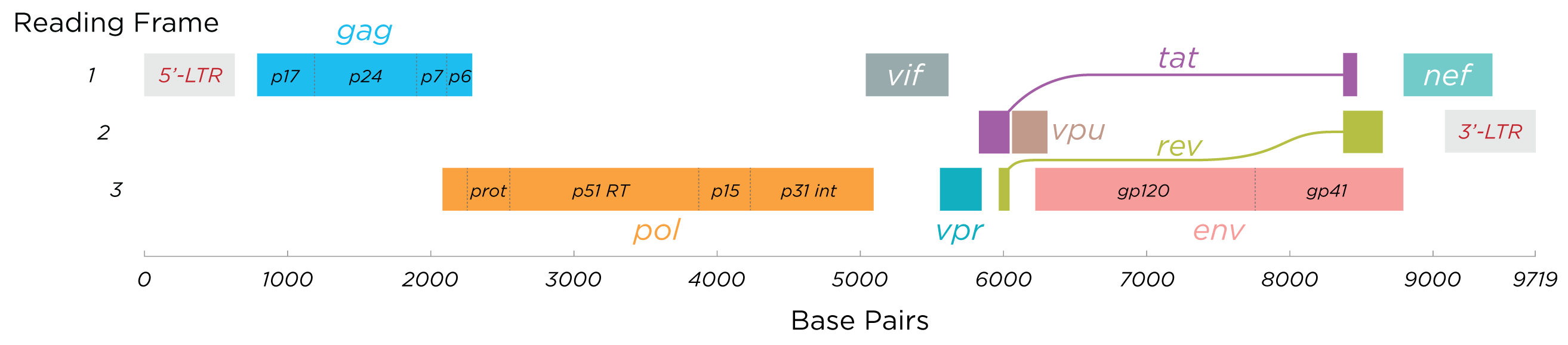
The complete HIV-1 genome has been solved to single-nucleotide resolution. Unlike other retroviruses, HIV encodes a small set of viral proteins that interact cooperatively with each other and with host proteins to hijack the cellular machinery. The virion is roughly 100 nm in diameter, with its interior consisting of a cone-shaped core that encloses two copies of positive-sense single-stranded RNA. Packaging two RNA copies (a state known as pseudodiploidy) facilitates recombination during reverse transcription, aids in template switching to bypass breaks in the RNA, and may serve structural roles in viral replication. The RNA is approximately 9,749 nucleotides long, featuring a 5’ cap, a 3’ poly(A) tail, and multiple open reading frames (ORFs) that encode both structural proteins and regulatory factors.
The single-stranded RNA is tightly bound to nucleocapsid proteins (p7) and is associated with other components—such as the late assembly protein (p6), reverse transcriptase, and integrase—that are essential for virion formation. The envelope, derived from the host cell membrane and reinforced by the viral matrix (p17), carries a few copies of the glycoprotein spike. These spikes, heavily shielded by N-linked glycans (predominantly of the high-mannose type), are major targets for neutralizing antibodies and vaccine development.
Genome Organization
The HIV genome contains several major genes that encode structural proteins common to all retroviruses and accessory genes unique to HIV. In total, nine genes encode fifteen viral proteins. These proteins are initially produced as polyproteins that are later processed into:
- Gag: Group-specific antigen forming the virion’s core structure.
- Pol: Enzymes required for replication, including reverse transcriptase, RNase H, integrase, and protease.
- Env: Envelope glycoprotein precursor (gp160), which is processed into gp120 and gp41.
HIV also produces regulatory and accessory proteins via differential RNA splicing. There are three main types of transcripts:
- A 9.2 kb unspliced transcript encoding the gag and pol precursors.
- A 4.5 kb singly spliced transcript encoding env, Vif, Vpr, and Vpu.
- A 2 kb multiply spliced transcript encoding the regulatory proteins Tat, Rev, and Nef.
Proteins Encoded by the HIV Genome
| Class | Gene Name | Primary Protein Products | Processed Protein Products |
|---|---|---|---|
| Viral Structural Proteins | gag | Gag polyprotein | MA, CA, SP1, NC, SP2, P6 |
| Viral Structural Proteins | pol | Pol polyprotein | RT, RNase H, IN, PR |
| Viral Structural Proteins | env | gp160 | gp120, gp41 |
| Essential Regulatory Elements | tat | Tat | — |
| Essential Regulatory Elements | rev | Rev | — |
| Accessory Regulatory Proteins | nef | Nef | — |
| Accessory Regulatory Proteins | vpr | Vpr | — |
| Accessory Regulatory Proteins | vif | Vif | — |
| Accessory Regulatory Proteins | vpu | Vpu | — |
Viral Structural Proteins
The gag gene encodes a polyprotein that is cleaved by the viral protease into several components: the matrix (p17), the capsid (p24), spacer peptides (SP1/p2 and SP2/p1), the nucleocapsid (p7), and the P6 protein. The pol gene encodes enzymes vital for viral replication—including reverse transcriptase, RNase H, integrase, and protease—while the env gene encodes gp160, which is processed by host proteases into the surface glycoprotein gp120 and the transmembrane protein gp41.
Essential Regulatory Elements
The regulatory proteins Tat and Rev are crucial for HIV replication. Tat enhances transcription by binding to the TAR element near the 5′ LTR, and Rev facilitates the export of unspliced or singly spliced viral RNAs from the nucleus by interacting with the Rev response element (RRE).
Accessory Regulatory Proteins
Accessory proteins, including Nef, Vpr, Vif, and Vpu, assist in various functions during the viral life cycle. Vpr aids in the nuclear import of the preintegration complex and can cause G2 cell cycle arrest. Vif enhances the infectivity of viral particles in specific cell types. Nef increases viral infectivity and may induce apoptosis in host cells. Vpu is involved in CD4 degradation and promotes the release of mature virions. In some HIV-1 isolates, an additional gene, tev, exists as a fusion product of tat, env, and rev.
RNA Secondary Structure
The HIV RNA genome contains several conserved secondary structure elements that play important roles in the viral life cycle. The 5′ untranslated region (UTR) is organized into multiple stem-loop structures (including the TAR element, polyadenylation signal, primer binding site (PBS), dimerization initiation site (DIS), major splice donor, and the ψ packaging signal). Additional RNA structures, such as the gag stem loop 3 (GSL3) and a cis-regulatory stem loop located between the protease and reverse transcriptase genes, are thought to influence reverse transcription and RNA packaging.
V3 Loop
The V3 loop is a variable region of the gp120 envelope glycoprotein that is critical for determining the virus’s tropism. By interacting with chemokine receptors (typically CCR5 or CXCR4) on host cells, the V3 loop plays a pivotal role in viral entry. Its structure makes it a key target for both therapeutic interventions and vaccine development.